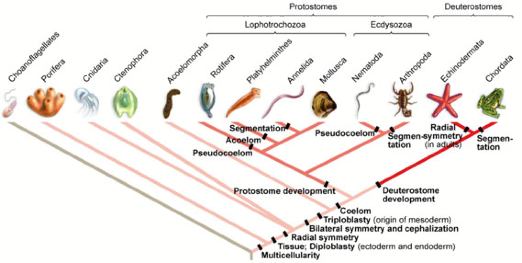
Analogous features are those that have the same function, but are not derived from the same things. The bacterial and the eukaryote flagella, for example. Homologous features are those that are derived from the same thing, but may or may not have the same function. Bird and insect wings, are an example,as are the different versions of Ubx.
Your talking off the topic Barbarian...
Then it was probably a mistake for you to bring up homology.
shrimp MNSYFEQN-GFYGSHP
fruitfly MNSYFEQASGFYG-H
mozzie MNSYFEQ-TGFYG-HP
worm MSSFFEPANMSNTIER
Just because the Ubx protein looks the same for all four organisms (the first 10 AA in the sequence) does not mean they have a common origin as evolution assumes.
As you learned, the differences in Ubx sort out in a way that confirms evolutionary inferences (infererences are from evidence, not assumptions) from other evidence. The differences in the sequence you posted shows fruitflies and mosquitos most closely related, than they to shrimp, with velvet worms most distantly related to them all.
Science doesn't deal in "proof", but likelihoods. This is yet another demonstration of common descent, consistent with evidence from genetics, fossil record, embyrology, and anatomy.
Regarding the question of whether or not a point mutation affects Ubx:
Depends. If it changes the shape of an active site, yes.
Good we agree, but how is a similar segment of a protein strand an indication of a theory of origins ?
They diverge over time, by mutation. We know this is a fact, because we can test it on organisms of known descent.
Suppose two Ubx strands were 90% the same, does that mean they must be of the same kind?
"Kind" is a religious belief, not a real taxon. We merely compare. So as you learned, the degree of similarity of genes demonstrated the same evolutionary relationships as shown by other evidence.
No, to me a single AA in the wrong place causes massive changes to the Ubx function, and sometimes it does not.
Told you. This is why we can trace phylogenies by the degree of change. The variable regions of the protein will change at a relatively constant rate, which, as you saw, shows the same phylogenies as several other lines of evidence.
Can you post the full AA sequence for the mozzie UbX, fruit fly Ubx and Worm Ubx and the Shrimp Ubx, so we can ascertain how similar they really are?
I don't know if that's available without a subscription to a journal. I'll take a look. I'll bet you a chocolate chip cookie that if we find some big molecules to compare between these taxa, the results will be the same. Bet?
The point is whether or not a single mutation will change the activity of Ubx. Many of them won't.
I agree.. point mutations won't change a Ubx function,
Some will.
so why use this AA pattern thing as a theory for origins ?
Because data from organisms of known descent shows that the degree of difference is a good measure of relatedness.
If not, unlikely to cause any change of function. There are numerous homologues of cytochrome C, for example, from bacteria to animals, and it seems to all work the same; essentially interchangeable.
The differences are predicted by evolutionary theory.
Nope. Demonstrable in organisms of known descent.
how does AA sequence in four organism predict origins ?
As you learned, the differences show the same degrees of relatedness as other sources of evidence.
Go back and look at the entire strand you posted. The phylogeny derived from your evidence (number of differences) shows the two insects to be most closely related,
OK the fly and mozzie have 30% of the Ubx AA sequences the same, how does that show they are the same in origins
Because they are more like each other than either is to the other organisms.
the insects and the crustacean to be more closely related to each other than any is to the velvet worm. Precisely what evolutonary theory predicts.
Creation theory would predict that too.
Nope. In fact, creationists used to deny this evidence:
In July 1983, the Public Broadcasting System televised an hour-long program on creationism. One of the scientists interviewed, biochemist Russell Doolittle, discussed the similarities between human proteins and chimpanzee proteins. In many cases, corresponding human and chimpanzee proteins are identical, and, in others, they differ by only a few amino acids. This strongly suggests a common ancestry for humans and apes. Gish was asked to comment. He replied:
"If we look at certain proteins, yes, man then -- it can be assumed that man is more closely related to a chimpanzee than other things. But on the other hand, if you look at certain other proteins, you'll find that man is more closely related to a bullforg than he is to a chimapanzee. If you focus your attention on other proteins, you'll find that man is more closely related to a chicken than he is to a chimpanzee."
Duane Gish of the Institute for Creation Research, denying the fact. (he was completely wrong, of course)
http://www.holysmoke.org/gishlies.htm
The shorter the strand, the less you can determine. Hence your lower amount of evidence only shows arthropods alike to the exclusion of velvet worms, while longer strands also distinguish insects from crustaceans.
"Homologous" doesn't necessarily mean "identical." And in this case, they aren't.
Forget about being scientifically accurate in speech.
Imprecision in science is useless. Sorry.
Do you have all species of Ubx for flies, shrimp's and mozzies and worms to compare ?
Well, hexapoda, using a number of Hox domains:
Nicely fits phylogenies from other sources of evidence.
Are you saying fly Ubx are different between species ? How different ?
That would be a concern for Creation model because why would God create a Ubx for fruit flies only to have it change radically....unless of course it is designed to change, after all it is a protein, not a DNA sequence is it ?
Each sort is from a slightly different DNA sequence, of course. But you're bumping up against the truth. Life indeed was created to change and adapt to new environments. No magic; He did it all by natural means.
SO I am not sure what to expect because the Bible does not explain Creation on the genetics level.
Some things, He left for us to find out. It's why he gave you a mind. It's not a sin to use it.
Regarding the claim of creationist models of Ubx;
Many things that science has discovered quickly become claimed to be part of the "creation model", but only after the fact. It would be more convincing you could show us a prediction for Ubx before this was known.
(nothing)
One of the major frustrations for creationists, is their inability to explain why this should be so only in cases of homology, but not for analogous features.
Can you unpack this statement, I am not following you at all....use simple words....
Sure. As Gish admitted, the creationist model says that distantly-related (by evolutionary terms) organisms should often have more similar molecules than closely-related ones. But that's not the case. Creationists are quite unable to explain why Gish was so wrong.
Precisely what evolutionary theory predicts. Differences, but less with the order than within insecta. And less within insecta than within arthropodia.
Have you tested this theory?
Yep. Tested on populations of known descent, it works.
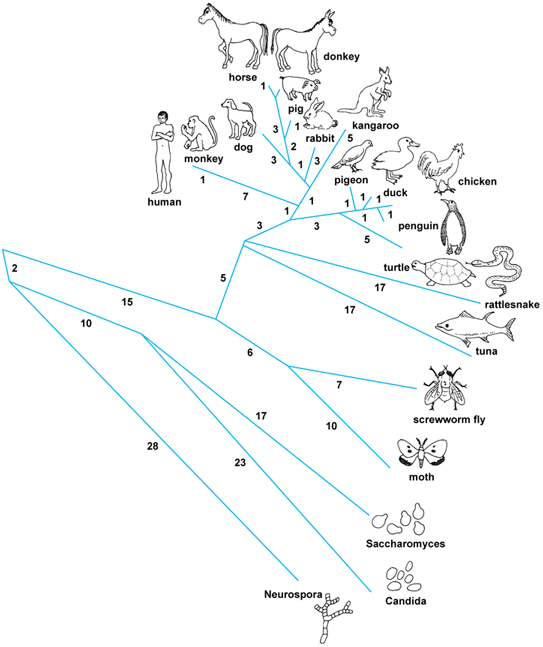
If we take each organism and count the number of differences between that organism and the others (as other scientists do, tracing human ancestry) we find that we get a distribution showing that the fly and the mosquito (both insects) are most closely related.
Well said Barbarian, I agree, but both Evolution and Creation origins predict this.
Nope. Notice Gish denies it.
Then the brine shrimp and the insects (all arthropods) are more closely related to each other, than any is to the velvet worm (an onychophoran).
Creation model would not agree, so this is a good test.
As you see, Ubx shows that to be the case. But let's see if we can find some other molecules to support the Ubx data:
Here's a phylogeny based on total DNA:
Here's one for cytochrome C:
\